Bristol University, UK
Gives you Wings?
Perhaps the most famous use of taurine in recent years has been in the energy drink Red Bull. Originating on college campuses and in nightclubs in Austria, the product quickly spread. The drink's marketing leans heavily on its unusual ingredient, and so integral is the substance to the brand that even the name derives from it - taurus is Latin for bull (as taurine was first isolated from ox bile in 1827). Red Bull promises that the taurine, along with caffeine and, in its original form, a great deal of sugar, will help to 'vitalise body and mind'.
 But what effect does the taurine in the drink actually have on the body? Research into this area is limited, but some researchers are sceptical [1]. They postulate that the effects of the drink can be attributed almost entirely to its caffeine content, and suggests that the increased effect compared to a cup of coffee (which contains a similar amount of caffeine) is only due to the temperature of the two drinks. A cold cup of coffee should have essentially the same physiological effect. Also they suggest a psychosomatic element to the drink's effects. In terms of the muscular effect of taurine, while it is possible that ingesting extra taurine would increase the force generation of the muscles no studies have been completed to demonstrate this and, given the naturally occurring levels of taurine in the body, it seems unlikely the dose added by Red Bull would cause a significant enough increase to have noticeable effects on the muscles.
But what effect does the taurine in the drink actually have on the body? Research into this area is limited, but some researchers are sceptical [1]. They postulate that the effects of the drink can be attributed almost entirely to its caffeine content, and suggests that the increased effect compared to a cup of coffee (which contains a similar amount of caffeine) is only due to the temperature of the two drinks. A cold cup of coffee should have essentially the same physiological effect. Also they suggest a psychosomatic element to the drink's effects. In terms of the muscular effect of taurine, while it is possible that ingesting extra taurine would increase the force generation of the muscles no studies have been completed to demonstrate this and, given the naturally occurring levels of taurine in the body, it seems unlikely the dose added by Red Bull would cause a significant enough increase to have noticeable effects on the muscles.
However, other researchers' findings support the company's claims. Seidl et al [2] report a double blind placebo controlled survey on a number of university students who demonstrated increased motor response and alertness compared to those receiving the placebo. Unfortunately the test did little to determine which of the drink's ingredients were responsible for the effects.
So what is taurine?
Although by the strictest definition, taurine is not an amino acid as it does not contain a carboxylic acid COOH group [3], it is generally referred to as one in published literature [4, 32,33,37]. At pH 7, the molecule exists as a zwitterion, in which the NH2 end of the molecule exists as NH3+ and the opposite end as SO3-. This gives the molecule two polar ends and a non-polar carbon chain centre, allowing for a great many possible binding interactions. Unusually for a biological compound, taurine does not exhibit chirality.
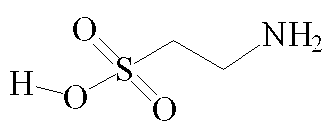
Taurine - (2-aminoethane sulphonic acid) [1]
Its Role in Biology and Pharmacy
Taurine is synthesised by the human body, primarily in the liver by oxidation followed by decarboxylation of the amino acid cysteine.
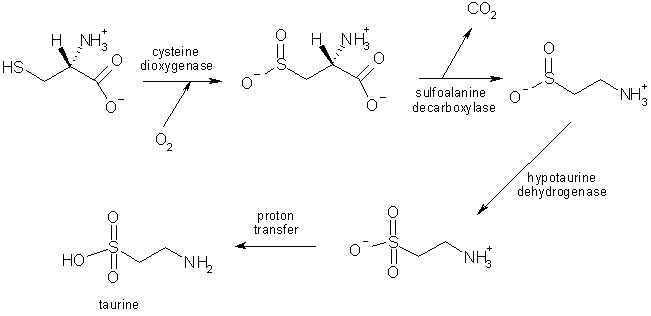
The biosynthesis of taurine from cysteine in the liver [5].
Taurine is one of a group of organic compounds which has been formed in experiments designed to simulate early-Earth conditions along with electrical discharges to simulate lightning. This result is often cited as evidence for the ease with which sulfur-containing biological molecules could be naturally synthesised under prebiotic conditions [6]. The conclusion is somewhat controversial, however, as the reaction occurred because of the presence of large concentrations of methane and ammonium hydroxide, the abundance of which on prebiotic Earth is questionable [7].
With a few exceptions, liver-synthesised taurine is not incorporated into polypeptides but is found free, lending it a unique set of properties, including a separate intercell transport mechanism and an independence from protein synthesis and catabolism [8]. Taurine has been linked to a wide range of bodily functions and Stapleton et al (1998) list roles in "osmoregulation, antioxidation, detoxification and stimulation of glycolysis and glycogenesis". The synthesis pathway of taurine is especially active in the early stages of life, and taurine is found in breast milk, suggesting that it is particularly vital at this stage. Taurine also plays a role in muscle contraction, where it enhances the ability of the muscles to generate force by catalysing the uptake and release of calcium ions [9].
Experiments on rats have also yielded evidence of a very substantial role in detoxification in the liver - Waters et al [10] report that high doses of taurine administered before or soon after ingestion of an overdose of paracetemol protected the livers of rats from hepatotoxcity and serious liver damage from the drugs. Although the idea has yet to progress to any form of human trial, the prophylactic and therapeutic benefits of the substance in cases of paracetemol overdose are promising .
 In fact, several areas of medical research are interested in taurine's pharmaceutical potential. For example, phase two clinical trials are currently underway at McLean Hospital in Massachusetts using taurine as an anti-manic agent to stabilise the mood of patients with bipolar disorder [11]. Tsuboyama-Kasaoka et al [12] also postulate a link between taurine deficiency and obesity in humans, showing that in mice, raising taurine levels in the body led to a greater resting rate of energy usage and less build-up of adipose tissue. The paper therefore suggests that taurine supplements may help prevent obesity.
In fact, several areas of medical research are interested in taurine's pharmaceutical potential. For example, phase two clinical trials are currently underway at McLean Hospital in Massachusetts using taurine as an anti-manic agent to stabilise the mood of patients with bipolar disorder [11]. Tsuboyama-Kasaoka et al [12] also postulate a link between taurine deficiency and obesity in humans, showing that in mice, raising taurine levels in the body led to a greater resting rate of energy usage and less build-up of adipose tissue. The paper therefore suggests that taurine supplements may help prevent obesity.
Taurine in animals
Taurine is necessary for normal skeletal muscle functioning in humans and mice [13]. Cats cannot synthesise taurine, and so need it added as supplements in their diets. Without it, they suffer conditions such as blindness, hair loss and tooth decay. Taurine is also essential in the early development of many types of perching birds. Parent birds with new offspring often seek out spiders (which are rich in taurines) with which to feed their young. so, in this case, taurine really does give them wings!
![]()
References
1. Kim W., "Debunking the Effects of Taurine in Red Bull Energy Drink", Nutrition Bytes, 9, (2003) 6.
2. Seidl R. et al, "A Taurine and Caffeine-containing drink stimulates cognitive performance and well-being", Amino Acids, 19, (2000) 635.
3. Sharp D., Dictionary of Chemistry (Abridged), 3rd Edition, Penguin Books, 2003.
4. Stapleton P. et. al, "Host defense - a role for the amino acid taurine?", J. Parenteral and Enteral Nutrition, 22, (1998) 42.
5. Moss G., "Taurine Biosynthesis", http://www.chem.qmul.ac.uk/iubmb/enzyme/reaction/misc/taurine.html, Queen Mary University of London, 1992.
6. Ferris J., "Chemical markers of Prebiotic Chemistry in Hydrothermal systems", in Origin of Life and Evolution of Biospheres V.22 Numbers 1-4, ed. N Holm, Springer, Netherlands, Jan. 1992, p.120.
7. Wigley T., Nature, 291, (1981) 213.
8. Chiarla C. et al, "The Relationship between Plasma Taurine and Other Amino Acid Levels in Human Sepsis", J. of Nutrition, 130, (2000) 2222.
9. K. Harrison, "Taurine", http://www.3dchem.com/molecules.asp?ID=22, (2002).
10. Waters E. et al, "Role of taurine in preventing acetaminophen-induced hepatic injury in the rat", Am. J. Physiol. Gastrointest. Liver Physiol., 280, (2001) 1274.
11. Murphy B. et al, "Taurine as an Anti-Manic Agent", (2005) http://www.clinicaltrials.gov/ct/show/NCT00217165
12. Tsuboyama-Kasaoka et al, "Taurine (2-Aminoethanesulfonic Acid) Deficiency Creates a Vicious Circle Promoting Obesity", Endocrinology, 147, (2006) 3276.
13. Wikipedia - taurine![]()